The Redwood Genome Project is a multi-year effort to sequence the coast redwood and giant sequoia genomes and lay the foundation for the development of genetic screening tools that can now be developed to aid in the conservation and management of these species.
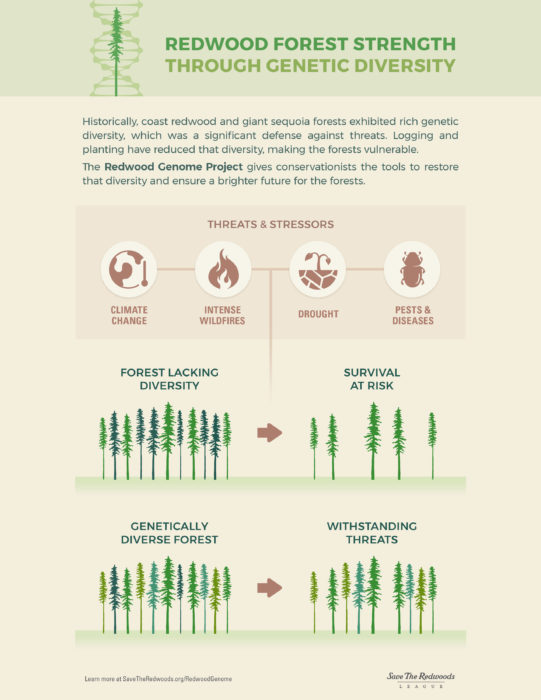
As the League seeks opportunities to restore coast redwood and giant sequoia forests—some of which have been damaged by decades of logging—we need to expand our understanding of how these species are adapting to their changing environments. What in the genetic code of the trees enables them to better withstand drought, wildfire, or climate change? How can we best achieve the resilience that genetic diversity can deliver? And, how can these lessons help guide the League’s important restoration work?
To answer these questions, the League in 2017 launched the Redwood Genome Project, a multi-year effort to sequence the coast redwood and giant sequoia genomes. Researchers from the League, University of California, Davis, Johns Hopkins University, University of Connecticut, and Northern Arizona University are leading the project.
Put simply, a genome is an organism’s complete set of genetic instructions, and is comprised of DNA, the unique chemical code that guides growth, development and health. Each genome contains all the information needed to build that organism and allow it to grow and develop.
New technologies made mapping the complex codes faster and more cost effective. Initial findings showed that the sequences for the coast redwood and giant sequoia are both quite complex, with the coast redwood genome is nearly nine times larger than the human genome.
This research helps lay the foundation for the development of genetic screening tools that can now be developed to inform management plans and help these species thrive for centuries to come.
Access the data
- Publication of coast redwood genome (December, 2021)
- Publication of giant sequoia genome (November, 2020)
- Sequence data available
Read more about the Redwood Genome Project at the links below.
PRESS RELEASE
- December 16, 2021 — Completed Redwood Genome Sequence Reveals Genes for Climate Adaptation and Offers Insights into Genetic Basis for Survival
For media inquiries, contact Robin Carr at (415) 766-0927 or redwoods@landispr.com.
PROJECT SUPPORT
We thank Ralph Eschenbach and Dr. Carol Joy Provan for their generous lead gift to support the Redwood Genome Project.
Tags: current, forest management, genetic diversity, genome sequencing, Johns Hopkins University, Northern Arizona University, redwood genome, UC Davis, University of California, University of Connecticut